Decomposition of the network
(in modules of predefined size)
For decomposing the network into submodules of a predefined size, use the parameter “-d” in heinz when running the script (see
http://www.mi.fu-berlin.de/w/LiSA/Heinz ).
All above steps are repeated in the same order. The difference is that the algorithm now not only searches for the optimal solution, but also for suboptimal solutions, excluding the proteins found in the optimal solution.
The same perl script can be used for generating each node attribute file for Cytoscape.
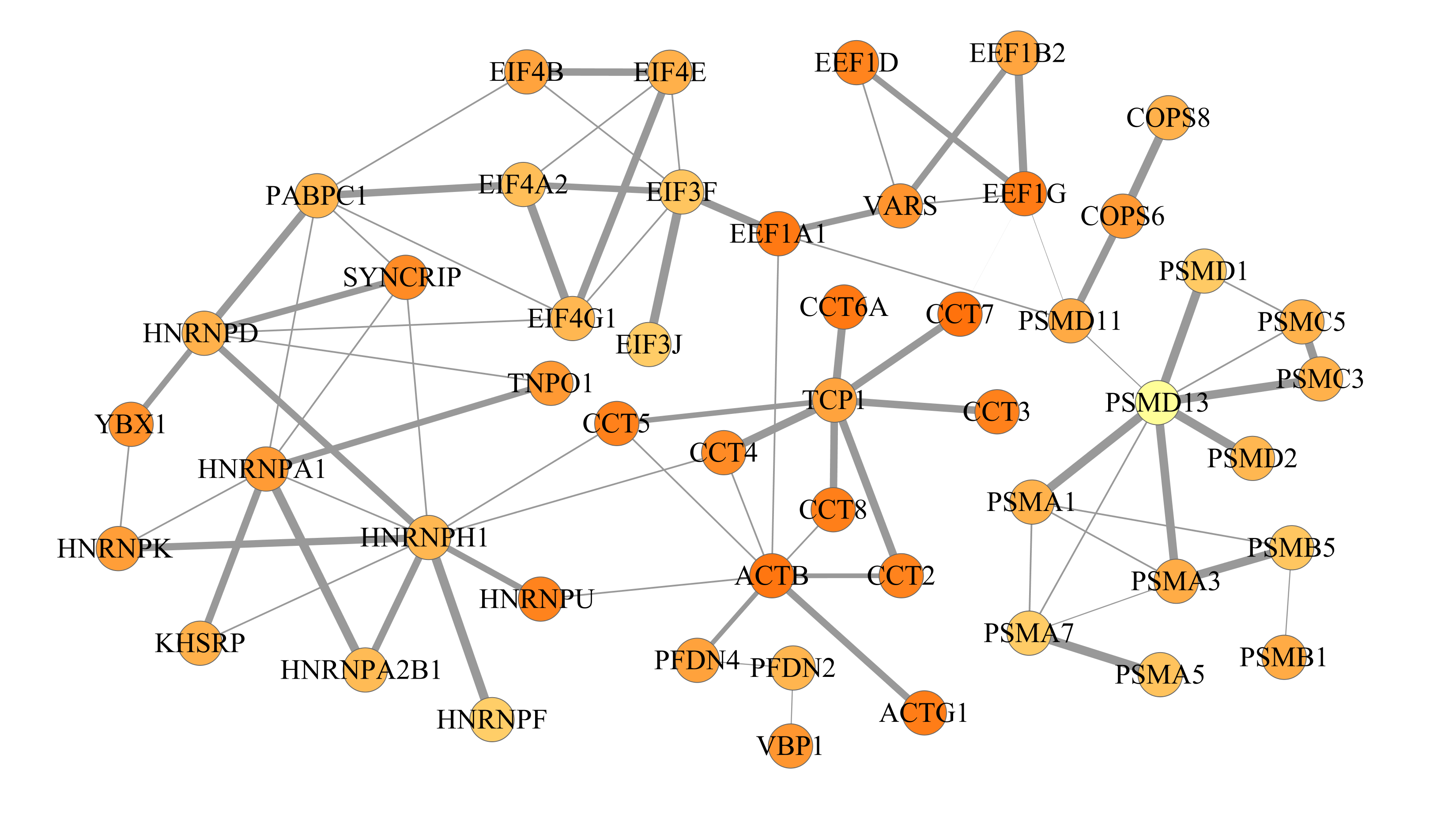
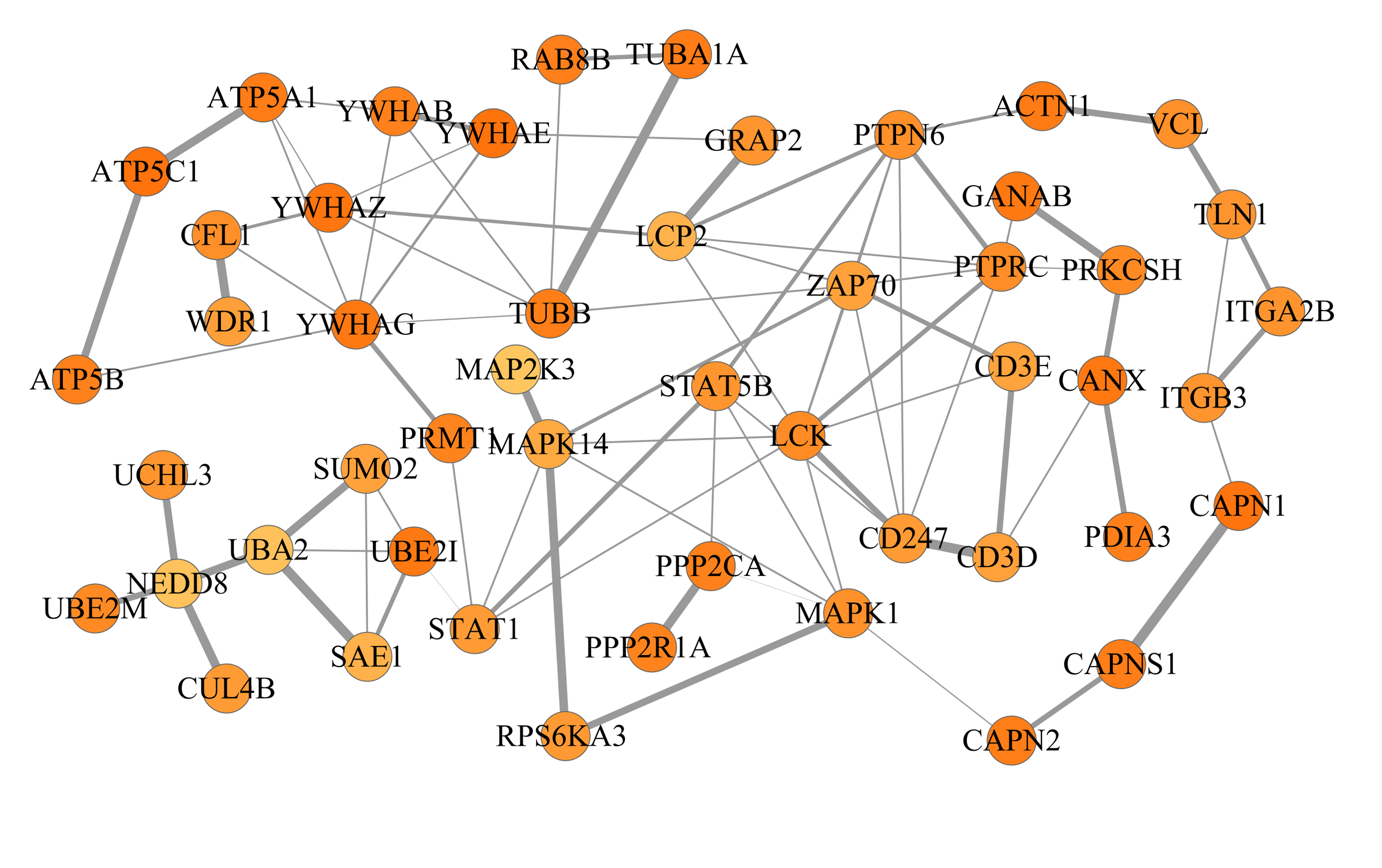
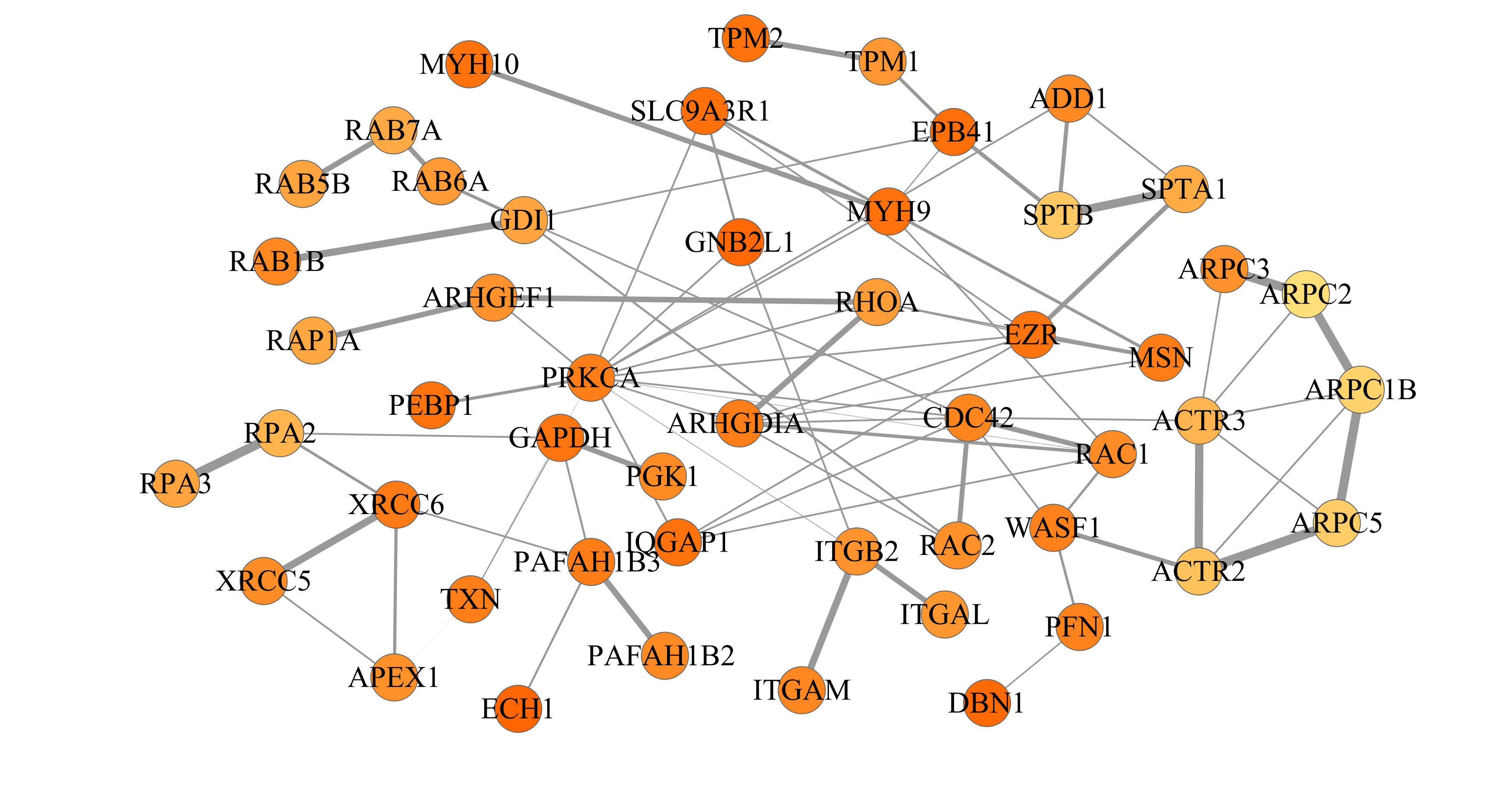
Here is an example from T-cells (example 2 above) after decomposing the network into 3 subnetworks of 50 proteins each:
heinz resulting edge files:
Solution 1 (download)
Solution 2 (download)
Solution 3 (download)
heinz resulting node files:
Solution 1 (download)
Solution 2 (download)
Solution 3 (download)
The final Cytoscape file can be downloaded here.
The final networks can be exported from Cytoscape and should look like this: