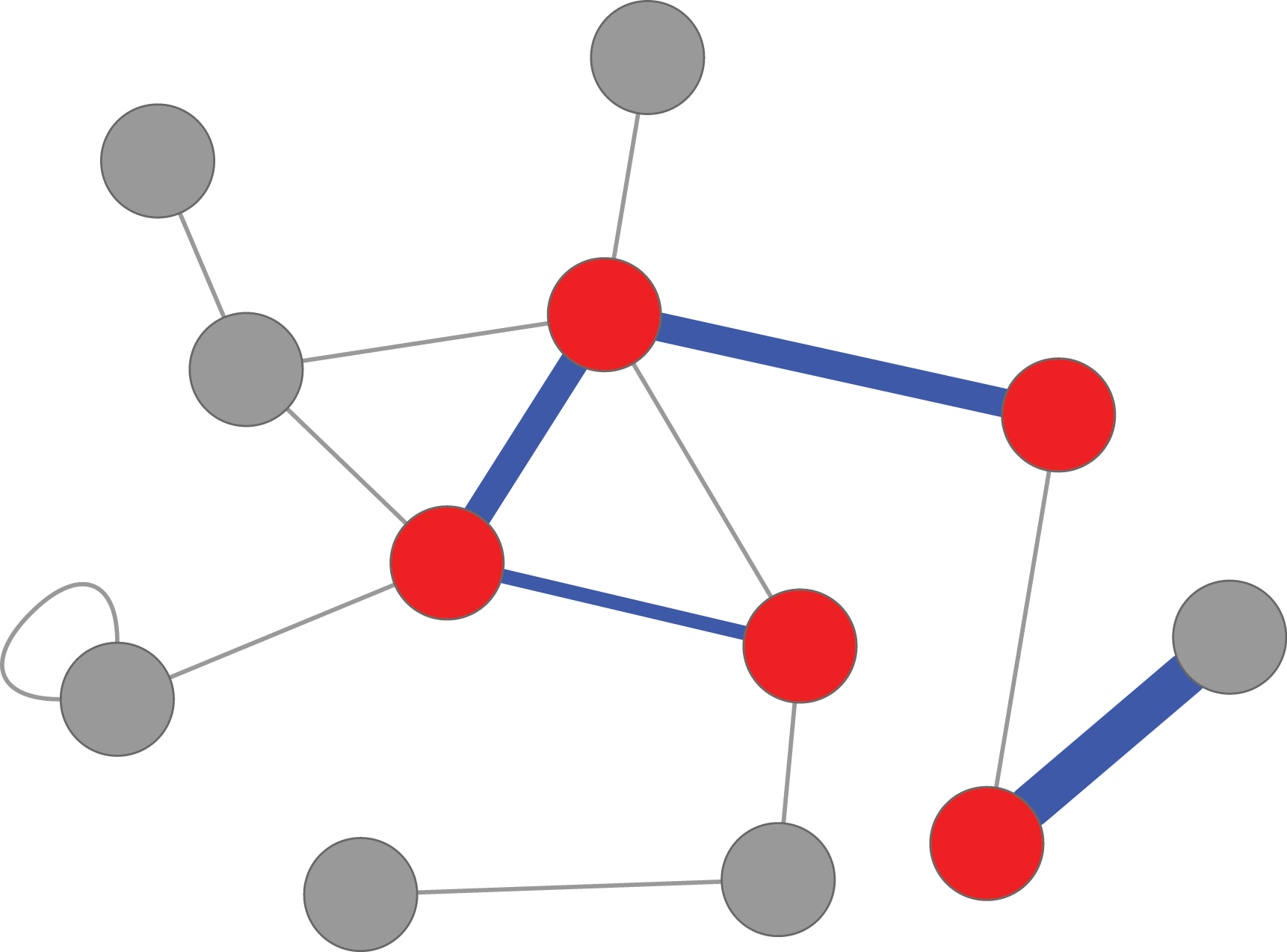
Heavy-induced subgraphs ("heinz") algorithm
- Introduction to heinz
The algorithm for identification of the optimal scoring subnetwork (heinz - heavy induced subnetworks) is based on the software dhea (district heating) from Ljubic et al. (2006). The C++ code is controlled over a Python script. The dhea code uses the commercial CPLEX callable library version 9.030 by ILOG, Inc. (Sunnyvale,CA). There is also free academic license available. A CPLEX library is needed for calculating the optimal solution.
The other routines, the dhea code and heinz.py Python script (current version 1.63) are publicly available for academic and research purposes within the heinz (heaviest induced subgraph) package of the open source library LiSA (http://www.mi.fu-berlin.de/w/LiSA/). The dhea code has to be positioned in the same folder as heinz.py to call the routine by the Python code. - Preparing files for heinz
heinz works with node (=protein) and edge (=interaction) files.
Node files contain protein scores and the file should always represent the data in the following way:#node score1 2 2.25349191365428 16 2.38788742334608 23 1.95476680990236
Edge files are functional interaction scores and the file should always represent the data in the following way:
#nodeA nodeB score1 2 259 -2.72455487068507 2 309 -2.78758900609407 2 348 -2.47279998651018
Examples: The files provided with the examples are ready for processing.
- heinz output files
a) edge file (= interactions file)
The algorithm searches for the maximum scoring subnetwork and provides an output file containing all edges in the interactome. The edges included in the solution are presented with their functional interaction scores, while all other edges obtain the value NaN as they were not chosen from the algorithm.
Example 1:
The final interaction file can be downloaded here.
Example 2:
The final interaction file can be downloaded here.
b) node file (= protein file)
The algorithm searches for the maximum scoring subnetwork and provides an output file containing all nodes in the network. The nodes included in the solution are presented with their scores, while all other nodes obtain the value NaN as they were not chosen from the algorithm.
Example 1:
The file can be downloaded here
Example 2:
The final protein file can be downloaded here.